[1] 8Introduction to R
CMOR Lunch’n’Learn
8 March 2023
Ross Wilson
Getting Started
What is R? What is RStudio?
- R is a programming language designed to undertake statistical analysis
- RStudio is an Integrated Development Environment (IDE) for R
- An IDE is a piece of software including a text editor and other tools to make programming (in this case programming in R, specifically) easier
- You don’t need to use RStudio to use R, but it makes it a lot easier
- To use RStudio, you also need R installed and working
Why learn R?
- Powerful and extensible
- Pretty much any statistical analysis you want to do can be done in R
- There are a huge number of freely-available user-written packages that provide all sorts of additional functionality
- Reproducibility
- By writing R scripts (code) for your analyses, they can be easily checked/replicated/ updated/adapted, by yourself and others
- High-quality graphics
- R has a lot of plotting functions to help you produce publication-quality figures
Getting set up
- Install R
- https://cloud.R-project.org/
- Choose the appropriate version for your operating system and follow the instructions
- Install RStudio
Getting to know RStudio
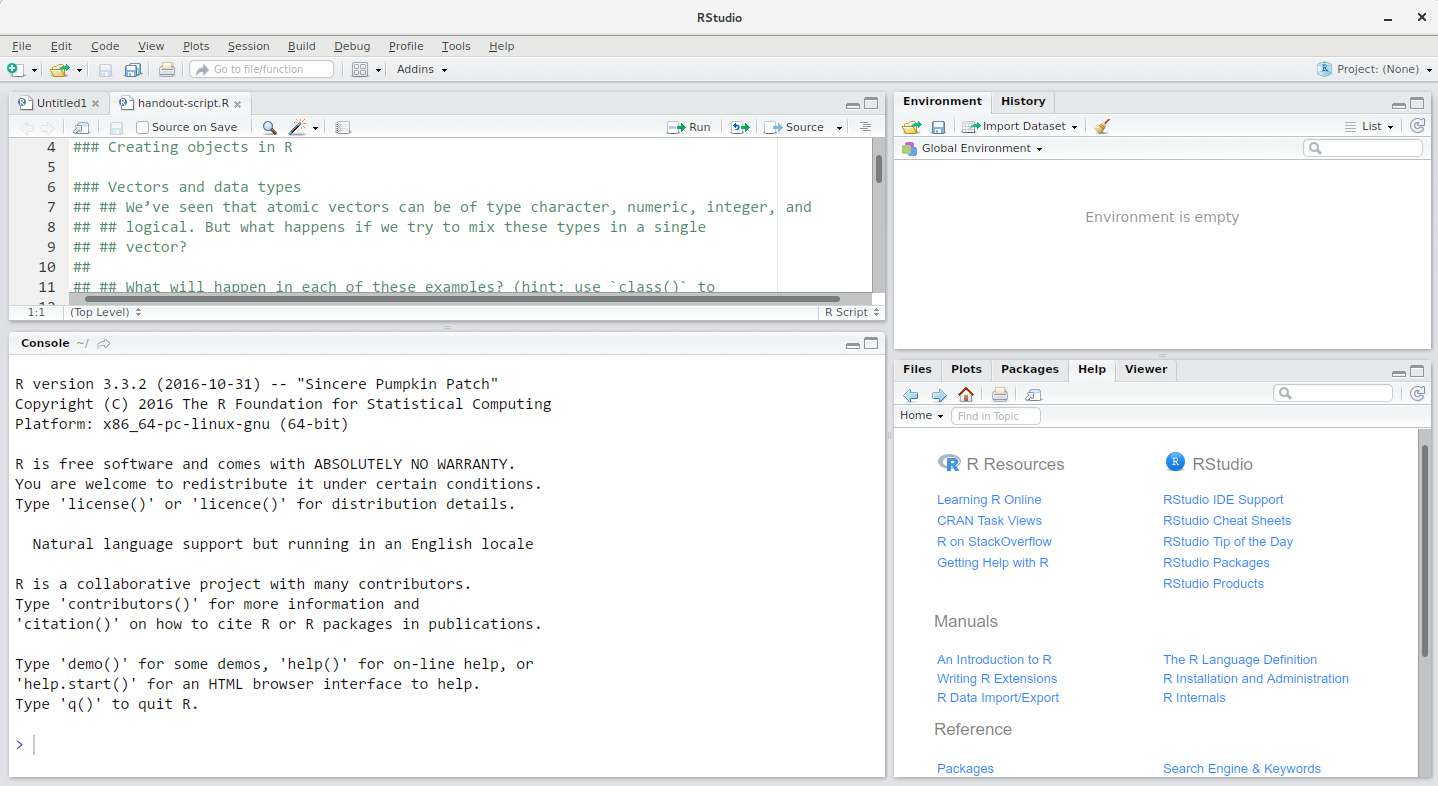
Source
Console
Environment/
History
Files/Plots/
Packages/
Help/Viewer
Starting with R
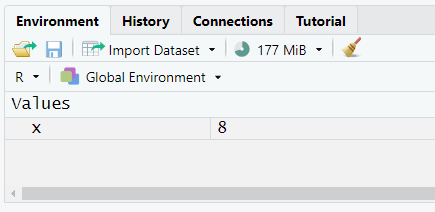
- We can type math in the console, and get an answer:
Starting with R
Functions
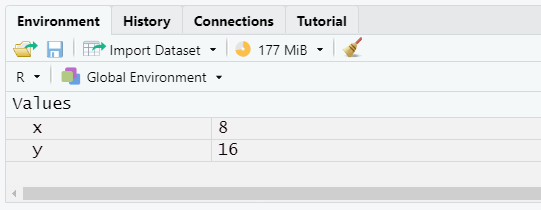
- Functions allow us to run commands other than simple arithmetic
- Functions consist of a set of input arguments, code that does something with those inputs, and a return value
Functions
- You can (and should!) also write your own functions
- Separating your code out into discrete functions makes it
- shorter
- easier to follow
- less error prone
Data types
- So far we have only seen single numeric values
- Data in R can take many forms
- The most basic data structure is the vector
Data types
- The basic data types are
numeric,character,logical(TRUEandFALSEvalues only), andinteger- Also
complexandraw, but we don’t need to worry about those
- Also
- In addition to vectors, more complex data structures include:
- lists: similar to vectors, but the elements can be anything (including other lists), and don’t need to all be the same
- matrices and arrays: like vectors, but with multiple dimensions
- data frames: more on these later
Subsetting
- We can extract values from within a vector (or other data structure) with square brackets
- We can use this in conjunction with a logical operator to do conditional subsetting
Working with Data Frames
Data frames
- A data frame is a tabular data structure (like a spreadsheet)
- Each column is a variable
- Each row is an observation
- Each column (variable) is a vector, so must contain a single data type
- But different columns can have different types
- We can read data from Excel or CSV spreadsheets, data files from Stata etc, previously saved R data frames, and much else…
The tidyverse
- The ‘
tidyverse’ is a collection of packages created by the company that makes RStudio - It contains a lot of functions designed to make working with data frames easier
tibble: a replacement for base data framesreadr: read tabular data like csv files (alsoreadxlfor Excel files,havenfor SPSS/Stata/SAS, and others for different file types)dplyr: data manipulationtidyr: reshaping and tidying dataggplot2: creating plotspurrr: functional programmingstringr: working with character stringsforcats: working with factor variables
Data frames
- Reading in a data frame with
read_csv():
# A tibble: 1,704 × 6
country year pop continent lifeExp gdpPercap
<chr> <dbl> <dbl> <chr> <dbl> <dbl>
1 Afghanistan 1952 8425333 Asia 28.8 779.
2 Afghanistan 1957 9240934 Asia 30.3 821.
3 Afghanistan 1962 10267083 Asia 32.0 853.
4 Afghanistan 1967 11537966 Asia 34.0 836.
5 Afghanistan 1972 13079460 Asia 36.1 740.
6 Afghanistan 1977 14880372 Asia 38.4 786.
7 Afghanistan 1982 12881816 Asia 39.9 978.
8 Afghanistan 1987 13867957 Asia 40.8 852.
9 Afghanistan 1992 16317921 Asia 41.7 649.
10 Afghanistan 1997 22227415 Asia 41.8 635.
# ℹ 1,694 more rowsManipulating data frames with dplyr
select()only a subset of variables
# A tibble: 1,704 × 3
year country gdpPercap
<dbl> <chr> <dbl>
1 1952 Afghanistan 779.
2 1957 Afghanistan 821.
3 1962 Afghanistan 853.
4 1967 Afghanistan 836.
5 1972 Afghanistan 740.
6 1977 Afghanistan 786.
7 1982 Afghanistan 978.
8 1987 Afghanistan 852.
9 1992 Afghanistan 649.
10 1997 Afghanistan 635.
# ℹ 1,694 more rowsManipulating data frames with dplyr
filter()only a subset of observations
# A tibble: 30 × 6
country year pop continent lifeExp gdpPercap
<chr> <dbl> <dbl> <chr> <dbl> <dbl>
1 Albania 2007 3600523 Europe 76.4 5937.
2 Austria 2007 8199783 Europe 79.8 36126.
3 Belgium 2007 10392226 Europe 79.4 33693.
4 Bosnia and Herzegovina 2007 4552198 Europe 74.9 7446.
5 Bulgaria 2007 7322858 Europe 73.0 10681.
6 Croatia 2007 4493312 Europe 75.7 14619.
7 Czech Republic 2007 10228744 Europe 76.5 22833.
8 Denmark 2007 5468120 Europe 78.3 35278.
9 Finland 2007 5238460 Europe 79.3 33207.
10 France 2007 61083916 Europe 80.7 30470.
# ℹ 20 more rowsManipulating data frames with dplyr
mutate()to create new variables
# A tibble: 1,704 × 7
country year pop continent lifeExp gdpPercap gdp_billion
<chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
1 Afghanistan 1952 8425333 Asia 28.8 779. 6.57
2 Afghanistan 1957 9240934 Asia 30.3 821. 7.59
3 Afghanistan 1962 10267083 Asia 32.0 853. 8.76
4 Afghanistan 1967 11537966 Asia 34.0 836. 9.65
5 Afghanistan 1972 13079460 Asia 36.1 740. 9.68
6 Afghanistan 1977 14880372 Asia 38.4 786. 11.7
7 Afghanistan 1982 12881816 Asia 39.9 978. 12.6
8 Afghanistan 1987 13867957 Asia 40.8 852. 11.8
9 Afghanistan 1992 16317921 Asia 41.7 649. 10.6
10 Afghanistan 1997 22227415 Asia 41.8 635. 14.1
# ℹ 1,694 more rowsManipulating data frames with dplyr
summarise()to calculate summary statistics
# A tibble: 1 × 1
mean_gdpPercap
<dbl>
1 7215.Manipulating data frames with dplyr
The power of
dplyris in combining several commands using ‘pipes’The previous command could be written:
Reshaping data frames
- Previously we said that data frames have variables in columns and observations in rows
- There may be different ways to interpret this in any given dataset
- Our dataset has country-by-year as the observation, and population, life expectancy, and GDP per capita as variables
- Sometimes it might make sense to have one row per country (observation), and multiple variables representing years
- These are known as ‘long’ and ‘wide’ format, respectively
Reshaping data frames
- The
tidyrpackage helps us transform our data from one shape to the otherpivot_wider()takes a long dataset and makes it widerpivot_longer()takes a wide dataset and makes it longer
- Recall our original dataset
# A tibble: 1,704 × 6
country year pop continent lifeExp gdpPercap
<chr> <dbl> <dbl> <chr> <dbl> <dbl>
1 Afghanistan 1952 8425333 Asia 28.8 779.
2 Afghanistan 1957 9240934 Asia 30.3 821.
3 Afghanistan 1962 10267083 Asia 32.0 853.
4 Afghanistan 1967 11537966 Asia 34.0 836.
5 Afghanistan 1972 13079460 Asia 36.1 740.
6 Afghanistan 1977 14880372 Asia 38.4 786.
7 Afghanistan 1982 12881816 Asia 39.9 978.
8 Afghanistan 1987 13867957 Asia 40.8 852.
9 Afghanistan 1992 16317921 Asia 41.7 649.
10 Afghanistan 1997 22227415 Asia 41.8 635.
# ℹ 1,694 more rowsReshaping data frames
- We can reshape this to be wider (one row per country)
gapminder_wide <- gapminder %>%
pivot_wider(id_cols = c(country, continent),
names_from = year, values_from = c(pop, lifeExp, gdpPercap))
gapminder_wide# A tibble: 142 × 38
country continent pop_1952 pop_1957 pop_1962 pop_1967 pop_1972 pop_1977
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Afghanistan Asia 8425333 9240934 10267083 11537966 13079460 14880372
2 Albania Europe 1282697 1476505 1728137 1984060 2263554 2509048
3 Algeria Africa 9279525 10270856 11000948 12760499 14760787 17152804
4 Angola Africa 4232095 4561361 4826015 5247469 5894858 6162675
5 Argentina Americas 17876956 19610538 21283783 22934225 24779799 26983828
6 Australia Oceania 8691212 9712569 10794968 11872264 13177000 14074100
7 Austria Europe 6927772 6965860 7129864 7376998 7544201 7568430
8 Bahrain Asia 120447 138655 171863 202182 230800 297410
9 Bangladesh Asia 46886859 51365468 56839289 62821884 70759295 80428306
10 Belgium Europe 8730405 8989111 9218400 9556500 9709100 9821800
# ℹ 132 more rows
# ℹ 30 more variables: pop_1982 <dbl>, pop_1987 <dbl>, pop_1992 <dbl>,
# pop_1997 <dbl>, pop_2002 <dbl>, pop_2007 <dbl>, lifeExp_1952 <dbl>,
# lifeExp_1957 <dbl>, lifeExp_1962 <dbl>, lifeExp_1967 <dbl>,
# lifeExp_1972 <dbl>, lifeExp_1977 <dbl>, lifeExp_1982 <dbl>,
# lifeExp_1987 <dbl>, lifeExp_1992 <dbl>, lifeExp_1997 <dbl>,
# lifeExp_2002 <dbl>, lifeExp_2007 <dbl>, gdpPercap_1952 <dbl>, …Reshaping data frames
- Often raw data will come in a wide format, and we want to reshape it longer for data analysis
gapminder_wide %>%
pivot_longer(pop_1952:gdpPercap_2007,
names_to = c(".value", "year"), names_sep = "_")# A tibble: 1,704 × 6
country continent year pop lifeExp gdpPercap
<chr> <chr> <chr> <dbl> <dbl> <dbl>
1 Afghanistan Asia 1952 8425333 28.8 779.
2 Afghanistan Asia 1957 9240934 30.3 821.
3 Afghanistan Asia 1962 10267083 32.0 853.
4 Afghanistan Asia 1967 11537966 34.0 836.
5 Afghanistan Asia 1972 13079460 36.1 740.
6 Afghanistan Asia 1977 14880372 38.4 786.
7 Afghanistan Asia 1982 12881816 39.9 978.
8 Afghanistan Asia 1987 13867957 40.8 852.
9 Afghanistan Asia 1992 16317921 41.7 649.
10 Afghanistan Asia 1997 22227415 41.8 635.
# ℹ 1,694 more rowsOther tidyverse packages
- We’ll come back to data visualisation with
ggplot2in a later session purrrprovides tools for functional programming- If we have several similar datasets, or subgroups within our dataset, we don’t want to write out/copy-and-paste our code separately for each one
- With
purrr, we can usemap()(and similar) to apply a function to multiple inputs and extract all of the outputs
stringrandforcatsare worth looking at if you need to work with string or factor variables – we won’t cover them here
Resources
- This material was adapted from the Data Carpentries’ ‘R for Social Scientists’ course (https://preview.carpentries.org/r-socialsci/index.html)
- Hands-On Programming with R (https://rstudio-education.github.io/hopr)
- An introduction to programming in R (for non-programmers!)
- R for Data Science (https://r4ds.hadley.nz)
- An excellent practical introduction to using the tidyverse
- Advanced R (https://adv-r.hadley.nz) and R Packages (https://r-pkgs.org)
- More advanced – good next steps once you’re a bit more comfortable using R
Data analysis in R (regression)
Everything in R is an object
- This includes your fitted regression models
- Workflow:
- Import/tidy/manipulate raw data
- Run regression or other models
- Use the created model objects
- print to console
- save to file
- extract coefficient estimates
- plot results
- export to excel/word
- etc.
Fitted model objects
R has evolved over time to fit a variety of different needs and use cases
This gives it great flexibility and ability to meet the needs of different users
But, the diversity of interfaces, data structures, implementation, and fitted model objects can be a challenge
- There is often more than one way to fit a given model
- What you’ve learned about one model implemented in a given package may not translate well to working with other functions/packages
We’ll cover some tools to help bridge that gap
Fitted model objects
A few common (but not universal) features:
Models are described by a
formula: e.g.y ~ x + zData are provided in a data frame (or, equivalently, tibble)
coef(),vcov(),summary()can be used to extract the coefficient estimates, variance covariance matrix, and to print a summary of the fitted model
The broom package
broomprovides several functions to convert fitted model objects to tidy tibblesFunctions:
tidy(): construct a tibble that summarises the statistical findings (coefficients, p-values, etc.)augment(): add new columns to the original data (predictions/fitted values, etc.)glance(): construct a one-row summary of the model (goodness-of-fit, etc.)
The package works with several model fitting functions from base R and commonly-used packages
- Some other packages may also implement their own methods to work with these functions
First example – linear regression
Linear regression models can be fitted with the
lm()function (in thestatspackage, part of base R)We’ll start by loading the gapminder dataset from the previous session:
First example – linear regression
- Use
?lmto find the documentation
Call:
lm(formula = lifeExp ~ gdpPercap + continent, data = gapminder)
Coefficients:
(Intercept) gdpPercap continentAmericas
4.789e+01 4.453e-04 1.359e+01
continentAsia continentEurope continentOceania
8.658e+00 1.757e+01 1.815e+01 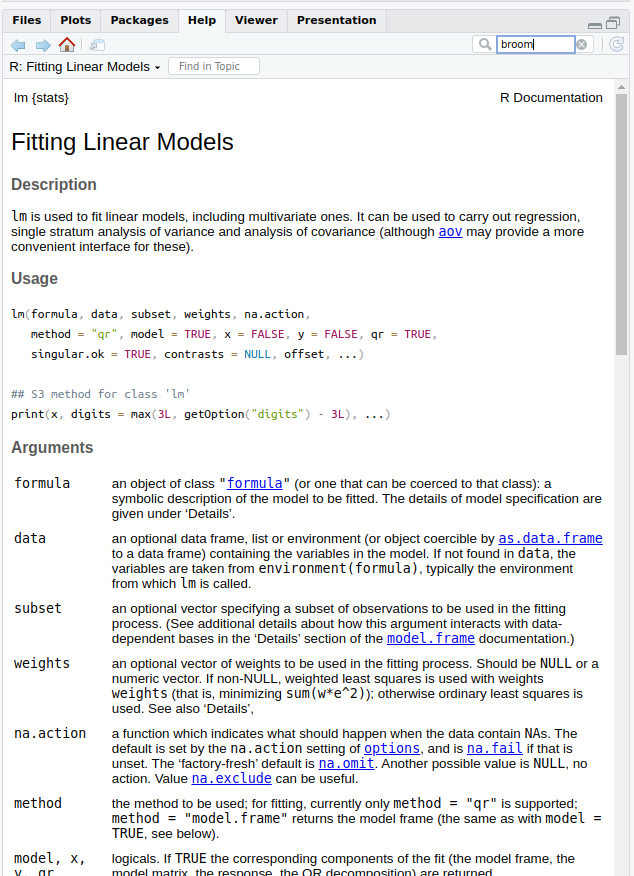
First example – linear regression
linear_regression_modelis now a fitted model objectIf we want, we can look at how this object is actually stored:
List of 13
$ coefficients : Named num [1:6] 4.79e+01 4.45e-04 1.36e+01 8.66 1.76e+01 ...
..- attr(*, "names")= chr [1:6] "(Intercept)" "gdpPercap" "continentAmericas" "continentAsia" ...
$ residuals : Named num [1:1704] -28.1 -26.6 -24.9 -22.9 -20.8 ...
..- attr(*, "names")= chr [1:1704] "1" "2" "3" "4" ...
$ effects : Named num [1:1704] -2455.1 311.1 100.1 -27.9 -223.1 ...
..- attr(*, "names")= chr [1:1704] "(Intercept)" "gdpPercap" "continentAmericas" "continentAsia" ...
$ rank : int 6
$ fitted.values: Named num [1:1704] 56.9 56.9 56.9 56.9 56.9 ...
..- attr(*, "names")= chr [1:1704] "1" "2" "3" "4" ...
$ assign : int [1:6] 0 1 2 2 2 2
$ qr :List of 5
..$ qr : num [1:1704, 1:6] -41.2795 0.0242 0.0242 0.0242 0.0242 ...
.. ..- attr(*, "dimnames")=List of 2
.. .. ..$ : chr [1:1704] "1" "2" "3" "4" ...
.. .. ..$ : chr [1:6] "(Intercept)" "gdpPercap" "continentAmericas" "continentAsia" ...
.. ..- attr(*, "assign")= int [1:6] 0 1 2 2 2 2
.. ..- attr(*, "contrasts")=List of 1
.. .. ..$ continent: chr "contr.treatment"
..$ qraux: num [1:6] 1.02 1.02 1.01 1.04 1.01 ...
..$ pivot: int [1:6] 1 2 3 4 5 6
..$ tol : num 1e-07
..$ rank : int 6
..- attr(*, "class")= chr "qr"
$ df.residual : int 1698
$ contrasts :List of 1
..$ continent: chr "contr.treatment"
$ xlevels :List of 1
..$ continent: chr [1:5] "Africa" "Americas" "Asia" "Europe" ...
$ call : language lm(formula = lifeExp ~ gdpPercap + continent, data = gapminder)
$ terms :Classes 'terms', 'formula' language lifeExp ~ gdpPercap + continent
.. ..- attr(*, "variables")= language list(lifeExp, gdpPercap, continent)
.. ..- attr(*, "factors")= int [1:3, 1:2] 0 1 0 0 0 1
.. .. ..- attr(*, "dimnames")=List of 2
.. .. .. ..$ : chr [1:3] "lifeExp" "gdpPercap" "continent"
.. .. .. ..$ : chr [1:2] "gdpPercap" "continent"
.. ..- attr(*, "term.labels")= chr [1:2] "gdpPercap" "continent"
.. ..- attr(*, "order")= int [1:2] 1 1
.. ..- attr(*, "intercept")= int 1
.. ..- attr(*, "response")= int 1
.. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
.. ..- attr(*, "predvars")= language list(lifeExp, gdpPercap, continent)
.. ..- attr(*, "dataClasses")= Named chr [1:3] "numeric" "numeric" "character"
.. .. ..- attr(*, "names")= chr [1:3] "lifeExp" "gdpPercap" "continent"
$ model :'data.frame': 1704 obs. of 3 variables:
..$ lifeExp : num [1:1704] 28.8 30.3 32 34 36.1 ...
..$ gdpPercap: num [1:1704] 779 821 853 836 740 ...
..$ continent: chr [1:1704] "Asia" "Asia" "Asia" "Asia" ...
..- attr(*, "terms")=Classes 'terms', 'formula' language lifeExp ~ gdpPercap + continent
.. .. ..- attr(*, "variables")= language list(lifeExp, gdpPercap, continent)
.. .. ..- attr(*, "factors")= int [1:3, 1:2] 0 1 0 0 0 1
.. .. .. ..- attr(*, "dimnames")=List of 2
.. .. .. .. ..$ : chr [1:3] "lifeExp" "gdpPercap" "continent"
.. .. .. .. ..$ : chr [1:2] "gdpPercap" "continent"
.. .. ..- attr(*, "term.labels")= chr [1:2] "gdpPercap" "continent"
.. .. ..- attr(*, "order")= int [1:2] 1 1
.. .. ..- attr(*, "intercept")= int 1
.. .. ..- attr(*, "response")= int 1
.. .. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
.. .. ..- attr(*, "predvars")= language list(lifeExp, gdpPercap, continent)
.. .. ..- attr(*, "dataClasses")= Named chr [1:3] "numeric" "numeric" "character"
.. .. .. ..- attr(*, "names")= chr [1:3] "lifeExp" "gdpPercap" "continent"
- attr(*, "class")= chr "lm"First example – linear regression
- More usefully, we can print a summary of the model
Call:
lm(formula = lifeExp ~ gdpPercap + continent, data = gapminder)
Residuals:
Min 1Q Median 3Q Max
-49.241 -4.479 0.347 5.105 25.138
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 4.789e+01 3.398e-01 140.93 <2e-16 ***
gdpPercap 4.453e-04 2.350e-05 18.95 <2e-16 ***
continentAmericas 1.359e+01 6.008e-01 22.62 <2e-16 ***
continentAsia 8.658e+00 5.555e-01 15.59 <2e-16 ***
continentEurope 1.757e+01 6.257e-01 28.08 <2e-16 ***
continentOceania 1.815e+01 1.787e+00 10.15 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 8.39 on 1698 degrees of freedom
Multiple R-squared: 0.5793, Adjusted R-squared: 0.5781
F-statistic: 467.7 on 5 and 1698 DF, p-value: < 2.2e-16First example – linear regression
- If we want to work with the results or combine/compare them with other models,
tidy()from thebroompackage will put them into a nice tidy tibble
# A tibble: 6 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 47.9 0.340 141. 0
2 gdpPercap 0.000445 0.0000235 18.9 8.55e- 73
3 continentAmericas 13.6 0.601 22.6 2.82e- 99
4 continentAsia 8.66 0.555 15.6 2.72e- 51
5 continentEurope 17.6 0.626 28.1 7.60e-143
6 continentOceania 18.1 1.79 10.2 1.50e- 23- And
glance()gives us several overall model statistics
# A tibble: 1 × 12
r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC deviance
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 0.579 0.578 8.39 468. 4.26e-316 5 -6039. 12093. 12131. 119528.
# ℹ 2 more variables: df.residual <int>, nobs <int>First example – linear regression
- The
lmtestandcarpackages provide tools for inference & hypothesis testing - Various clustered and heteroskedasticity-robust standard errors are provided by
sandwich- These work well together (and with
tidy()frombroom)
- These work well together (and with
library(lmtest)
library(sandwich)
linear_regression_model %>%
coeftest(vcov. = vcovCL, cluster = ~country) %>%
tidy()# A tibble: 6 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 47.9 0.904 53.0 0
2 gdpPercap 0.000445 0.000159 2.81 5.05e- 3
3 continentAmericas 13.6 1.56 8.69 8.32e-18
4 continentAsia 8.66 1.55 5.57 2.98e- 8
5 continentEurope 17.6 2.18 8.06 1.44e-15
6 continentOceania 18.1 2.77 6.55 7.36e-11Generalised linear regression
- Can be fitted with the
glm()function
(also in thestatspackage)
?glm
gen_linear_regression_model <- glm(lifeExp ~ gdpPercap + continent,
family = "quasipoisson", gapminder)
gen_linear_regression_model
Call: glm(formula = lifeExp ~ gdpPercap + continent, family = "quasipoisson",
data = gapminder)
Coefficients:
(Intercept) gdpPercap continentAmericas
3.875e+00 6.156e-06 2.490e-01
continentAsia continentEurope continentOceania
1.671e-01 3.092e-01 3.177e-01
Degrees of Freedom: 1703 Total (i.e. Null); 1698 Residual
Null Deviance: 4946
Residual Deviance: 2229 AIC: NA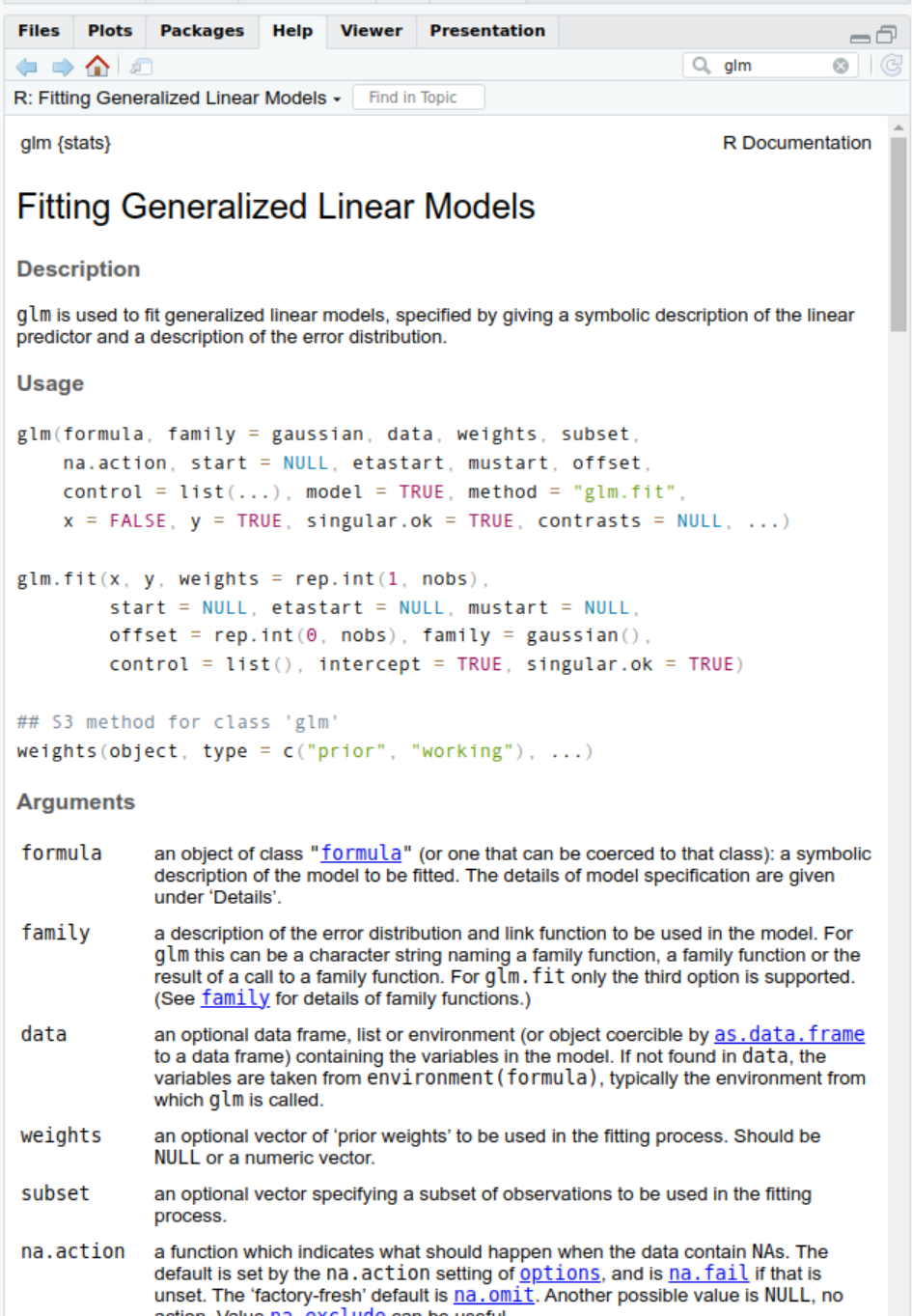
Generalised linear regression
- Again, we can print a summary of the model
Call:
glm(formula = lifeExp ~ gdpPercap + continent, family = "quasipoisson",
data = gapminder)
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.875e+00 6.524e-03 594.03 <2e-16 ***
gdpPercap 6.156e-06 3.475e-07 17.72 <2e-16 ***
continentAmericas 2.490e-01 1.054e-02 23.62 <2e-16 ***
continentAsia 1.671e-01 1.009e-02 16.55 <2e-16 ***
continentEurope 3.092e-01 1.054e-02 29.33 <2e-16 ***
continentOceania 3.177e-01 2.815e-02 11.29 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasipoisson family taken to be 1.279258)
Null deviance: 4945.6 on 1703 degrees of freedom
Residual deviance: 2229.4 on 1698 degrees of freedom
AIC: NA
Number of Fisher Scoring iterations: 4Generalised linear regression
- And use
tidy()andglance()from thebroompackage to produce nice tidy tibbles
# A tibble: 6 × 7
term estimate std.error statistic p.value conf.low conf.high
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 48.2 0.00652 594. 0 47.6 48.8
2 gdpPercap 1.00 0.000000347 17.7 1.42e- 64 1.00 1.00
3 continentAmericas 1.28 0.0105 23.6 6.86e-107 1.26 1.31
4 continentAsia 1.18 0.0101 16.6 3.57e- 57 1.16 1.21
5 continentEurope 1.36 0.0105 29.3 2.32e-153 1.33 1.39
6 continentOceania 1.37 0.0282 11.3 1.57e- 28 1.30 1.45What else?
There are many (many!) other packages that estimate different regression models or provide tools for diagnostic testing, post-hoc analysis, or visualisation of regression models
For example, the Econometrics Task View on cran.r-project.org
Data visualisation
Data visualisation with ggplot2
- There are a lot of tools available to create plots in R
ggplot2is the most well-developed and widely used
- We generally want data in long format for plotting
- One column for each variable
- One row for each observation
- We’ll use the
gapminderdataset from the previous session
The grammar of graphics
- dataset – self-explanatory
- geom – the geometric object used to represent the data
- mappings – which features of the geom represent which variables in the data
- stats – transformations of the data before plotting
- position – to avoid overplotting data points
- coordinate system – how the x and y axes are plotted
- faceting scheme – split the plot by subgroups
Data visualisation with ggplot2
- That’s the theory
- In practice, the easiest way is to build the plot up step-by-step (trial-and-error)
- start with the basic
ggplotobject
- start with the basic
- you can specify (some of) the mappings at this stage
Data analysis workflows and project organisation
Reproducible research
- What is ‘reproducibility’?
- Focus here is on computational reproducibility – can your results be replicated by someone (or yourself!) with access to your data, code, etc.
- Why might research not be reproducible?
- Raw data are not available or have been changed
- Intermediate steps taken to extract, clean, reshape, merge, or analyse data are not adequately recorded or described
- Software tools have changed or are no longer available since the analysis was conducted
- Errors in manually transcribing results from analysis software to final report (manuscript, etc.)
Reproducible research
Computers are now essential in all branches of science, but most researchers are never taught the equivalent of basic lab skills for research computing. As a result, data can get lost, analyses can take much longer than necessary, and researchers are limited in how effectively they can work with software and data. Computing workflows need to follow the same practices as lab projects and notebooks, with organized data, documented steps, and the project structured for reproducibility, but researchers new to computing often don’t know where to start.
— Wilson G, Bryan J, Cranston K, et al. Good enough practices in scientific computing. PLoS Comput Biol 2017;13:e1005510
Reproducible research
- Ensuring reproducibility (for others) can also have great benefits for you as the analyst/author
- When you come back to an analysis later, you know exactly what you did, why you did it, and how to replicate it if needed
- If data changes, errors are identified, or new analyses need to be conducted, your analysis and results can easily be updated
- The code and methods used in one project/analysis can be re-used in other work
- Keeping to a simple, standardised workflow for all projects allows you to switch easily & quickly between projects
- And prevents you having to make a whole bunch of new decisions every time you start something new
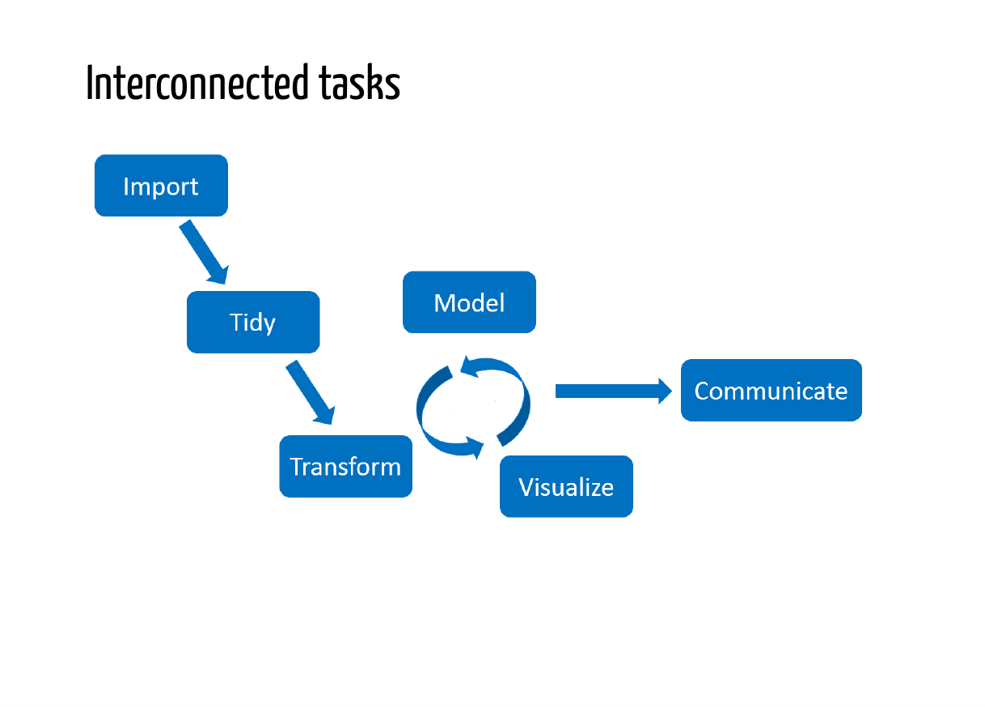
Key principles
- Raw data stays raw
- Source is real
- Design for collaboration
- (including with your future self)
- Consider version control
- Automation?
Key principles – Raw data
- Raw data stays raw
- Make raw data files read-only
Key principles – Raw data
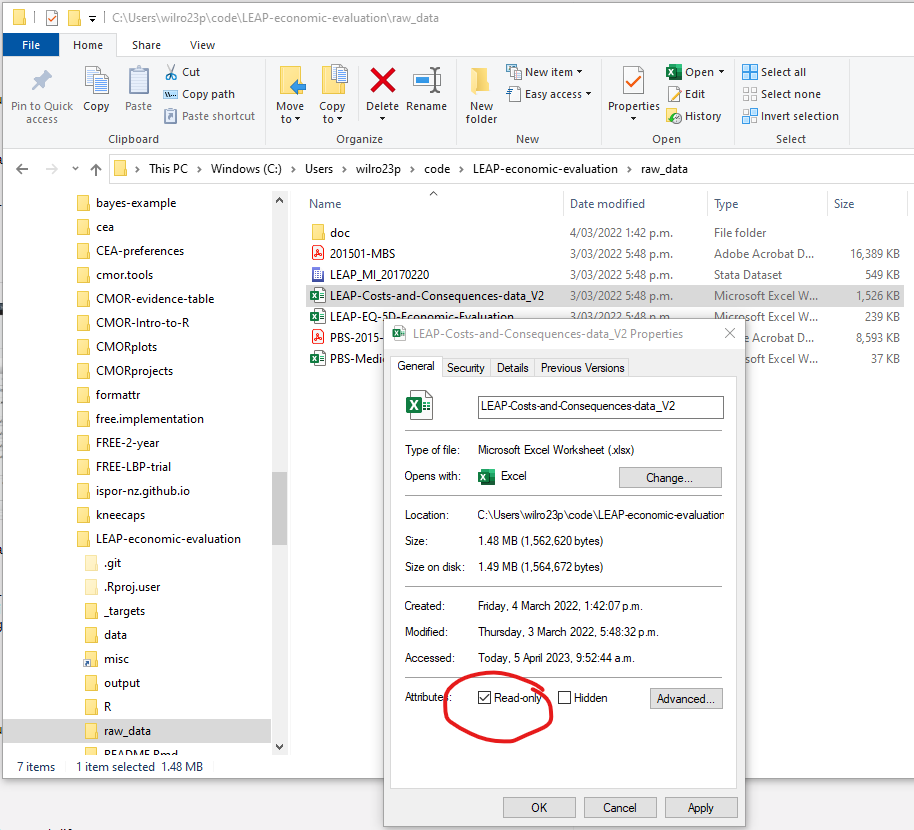
Key principles – Raw data
- Raw data stays raw
- Make raw data files read-only
- Can be relaxed (temporarily) if needed, but avoids accidental changes
- Most (probably all) changes to data should be scripted (in R, Stata, etc.) rather than made directly in raw data files
Key principles
- Raw data stays raw
- Source is real
- Design for collaboration
- (including with your future self)
- Consider version control
- Automation?
Key principles
- Raw data stays raw
- Source is real
- Design for collaboration
- (including with your future self)
- Consider version control
- Automation?
Key principles – Source files
- Source is real…
- …workspace is replaceable
- Any individual R session is disposable and can be replaced/recreated at any time, if you have raw data and appropriate source code
- Thinking in this way forces you to follow good reproducibility practices in your analysis workflow
Key principles – Source files
- What does ‘Source is real’ mean?
- Data import, cleaning, reshaping, wrangling, and analysis should all be conducted via script files
- or at least documented in thorough, step-by-step detail
- All source code should be saved (regularly!) so all of these steps can be reproduced at any time
- Important (or time-consuming) intermediate objects (cleaned datasets, figures, etc) should be explicitly saved, individually, to files (by script, not the mouse, if possible)
- Data import, cleaning, reshaping, wrangling, and analysis should all be conducted via script files
Key principles – Source files
- Start R with a blank slate, and restart often
Key principles – Source files
Key principles – Source files
- Start R with a blank slate, and restart often
- When you are running code interactively to try things out, you will be adding objects to the workspace, loading packages, etc
- If you inadvertently rely on these objects, packages, etc in your source files, you may not be able to reproduce your results in a new session later
- If you have saved source code and intermediate objects to file as you go, it is quick and easy to restart R (Ctrl+Shift+F10, in RStudio) regularly to check whether everything still works as expected
- It is much better to find this out after a few minutes than after a whole day’s work!
Key principles
- Raw data stays raw
- Source is real
- Design for collaboration
- (including with your future self)
- Consider version control
- Automation?
Key principles
- Raw data stays raw
- Source is real
- Design for collaboration
- (including with your future self)
- Consider version control
- Automation?
Key principles – Design for collaboration
- An analysis directory like this makes it very difficult for anyone (including yourself later) to follow the steps required to reproduce your results
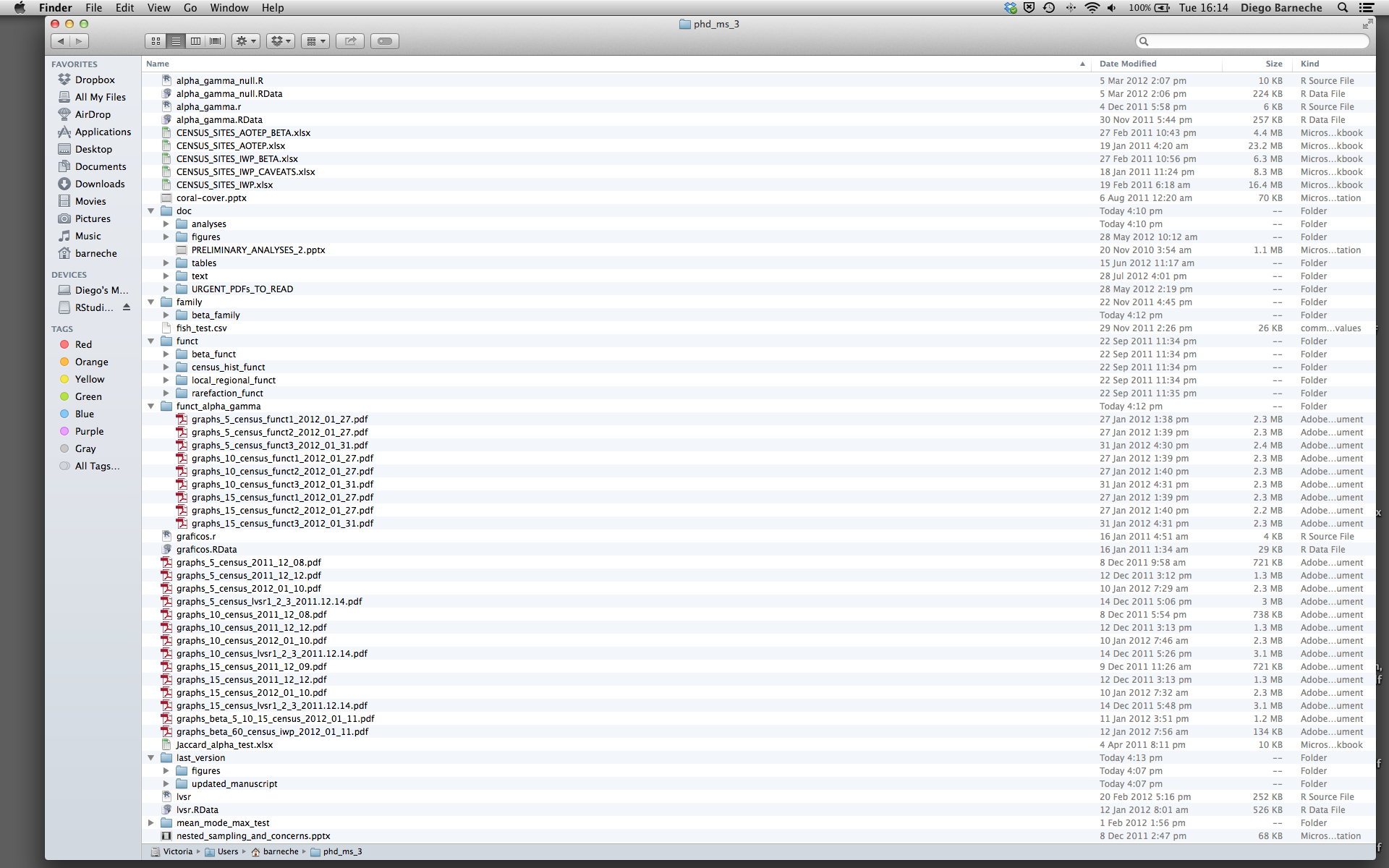
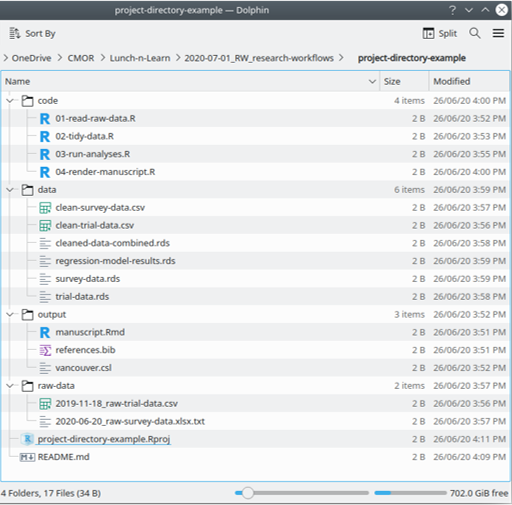
Key principles – Design for collaboration
- Much better to follow a logical structure (and the same across all of your projects)
- Set out high-level structure, e.g.
- read raw data
- tidy data for analysis
- run analyses on tidy data
- collate report/results of analyses
- Put raw data in its own directory (and read-only)
- Consider other structures/subfolders as appropriate for your project
- Set out high-level structure, e.g.
Key principles – Design for collaboration
- Break up complicated or repeated analysis steps into discrete functions
- Give functions and variables meaningful names
- Use comments liberally
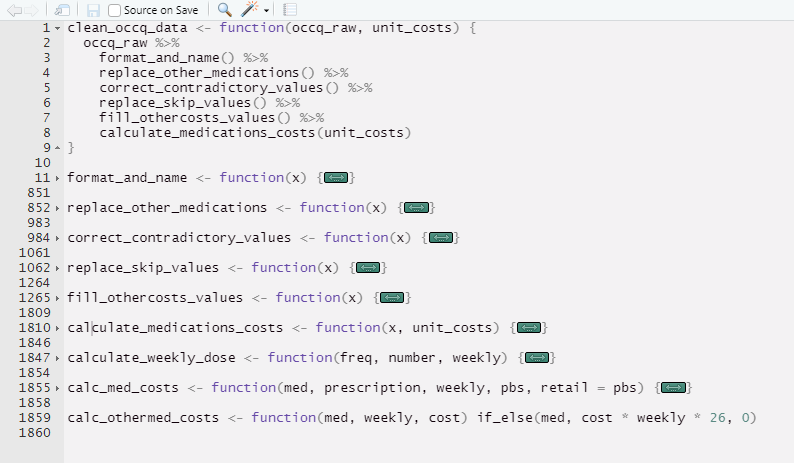
Key principles
- Raw data stays raw
- Source is real
- Design for collaboration
- (including with your future self)
- Consider version control
- Automation?
Key principles
- Raw data stays raw
- Source is real
- Design for collaboration
- (including with your future self)
- Consider version control
- Automation?
Key principles – Version control
Keeping track of changes to data and code (and being able to revert to a previous version if things go wrong) is critical for reproducible research
This is particularly true when collaborating with others
The best way to do this is with a version control system such as Git
Key principles – Version control
- We won’t go into this in detail today, but a quick Getting Started guide:
- Register a GitHub account (https://github.com)
- Install Git (https://git-scm.com) and complete basic setup
- Get a Git client (GUI)
- GitKraken is a good choice (https://gitkraken.com)
- GitHub offers a free client, GitHub Desktop (https://desktop.github.com/)
- RStudio has a basic Git client built-in, which is fine for much day-to-day use
- See https://happygitwithr.com/ for more detailed instructions
Key principles – Version control
- Using Git (& GitHub) in your projects
- A couple of ways to get this set up. Probably the easiest is:
- Create a project repository (repo) on GitHub
- Import (‘clone’) the repo as a new RStudio project
- Make changes as usual in RStudio (or any other program)
- ‘Commit’ (take a snapshot of the current state of the project) often to your local repo
- Less often (maybe once/day) ‘push’ (sync) the local repo to GitHub
- What to put under version control
- Source code
- Raw data (unless its very large)
What not to put under version control
- Most intermediate objects
- Miscellaneous supporting documents (pdf files, etc.)
- Final output in the form of word documents, etc.
Key principles
- Raw data stays raw
- Source is real
- Design for collaboration
- (including with your future self)
- Consider version control
- Automation?
Key principles
- Raw data stays raw
- Source is real
- Design for collaboration
- (including with your future self)
- Consider version control
- Automation?
Key principles – Automation
- Just as we treat source code as the ‘true’, reproducible record of each step of the analysis, we can record (and automate) how the sequence of individual steps fits together to produce our final results
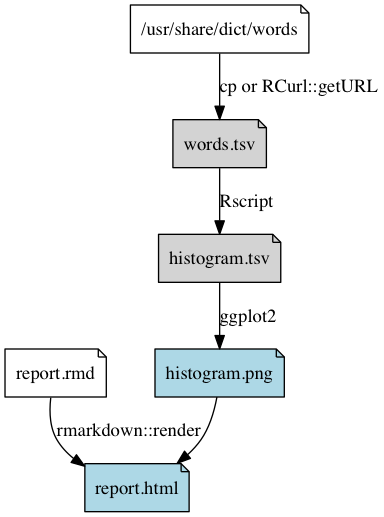
Can be as simple as something like:
Shows in what order to run the scripts, and allows us to resume from the middle (if, for example, you have only changed the file
02-create-histogram.R, there is no need to redo the first two steps, but we do need to rerun thecreate-histogramandrender-reportsteps- Each script should load the required inputs and save the resulting output to file
Key principles – Automation
For more complicated (or long-running) analyses, we may want to explicitly specify dependencies and let the computer figure out how to get everything up-to-date
Two good tools for doing this:
- make
- targets
Advantages of an automated pipeline like this:
- When you modify one stage of the pipeline, you can re-run your analyses to get up-to-date final results, with a single click/command
- Only the things that need to be updated will be re-run, saving time (important if some of your analyses take a long time to run)
Key principles – Automation
Make is a system tool, designed for use in software development, to specify targets, commands, and dependencies between files and selectively re-run commands when dependencies change
A Makefile is a plain text file specifying a list of these targets (intermediate/output files in the analysis workflow), commands (to create the targets), and dependencies (input files needed for each command)
words.txt: /usr/share/dict/words
cp /usr/share/dict/words words.txt
histogram.tsv: histogram.r words.txt
Rscript $<
histogram.png: histogram.tsv
Rscript -e 'library(ggplot2); qplot(Length, Freq, data=read.delim("$<")); ggsave("$@")'
report.html: report.rmd histogram.tsv histogram.png
Rscript -e 'rmarkdown::render("$<")'Key principles – Automation
targetsis an R package designed to do something very similar, but specifically designed for R projects
plan <- list(
tar_file(words, download_words()),
tar_target(frequency_table, summarise_word_lengths(words)),
tar_target(histogram, create_histogram(frequency_table)),
tar_file(report, render_report("reports/report.rmd", frequency_table, histogram))
)- Abstract workflow steps behind function calls (with meaningful names) as much as possible
- The plan should be a clear, high-level overview of the analysis steps required
Summary
- Treat raw data as read-only
- Save source, not the workspace
- Design for collaboration
- Follow a logical project structure
- Simplify code with human-readable abstractions (functions etc)
- Consider version control
- Consider automating your analyses

Summary
- Treat raw data as read-only
![]()
- Save source, not the workspace
![]()
- Design for collaboration
- Follow a logical project structure
![]()
- Simplify code with human-readable abstractions (functions etc)
![]()
- Follow a logical project structure
- Consider version control
- Consider automating your analyses
(if you are already writing
script files for your analyses)
(will get better
with practice)
(start with a simple high-level overview of the steps in the analysis,
even if you don’t want to do a full automated pipeline)
Resources
- Wilson G, Aruliah DA, Brown CT, Chue Hong NP, Davis M, et al. (2014) Best Practices for Scientific Computing. PLoS Biol 12(1):e1001745. https://doi.org/10.1371/journal.pbio.1001745
- Wilson G, Bryan J, Cranston K, Kitzes J, Nederbragt L, Teal TK (2017) Good enough practices in scientific computing. PloS Comput Biol 13(6):e1005510. https://doi.org/10.1371/journal.pcbi.1005510
- Bryan J, Hester J. What they forgot to teach you about R. https://rstats.wtf (especially Section I: A holistic workflow)
- Bryan J. STAT545 (based on the UBC course of the same name). https://stat545.com
- Bryan J. Happy Git and GitHub for the useR. https://happygitwithr.com
- For targets:
- McBain M. Benefits of a function-based diet (The {drake} post). https://milesmcbain.xyz/the-drake-post/
- Refers to the
drakepackage, which was a predecessor package oftargets. The same concepts all apply
- Refers to the
- Landau W. The {targets} R package user manual. https://books.ropensci.org/targets/
- McBain M. Benefits of a function-based diet (The {drake} post). https://milesmcbain.xyz/the-drake-post/
Writing reports in R
Data workflow
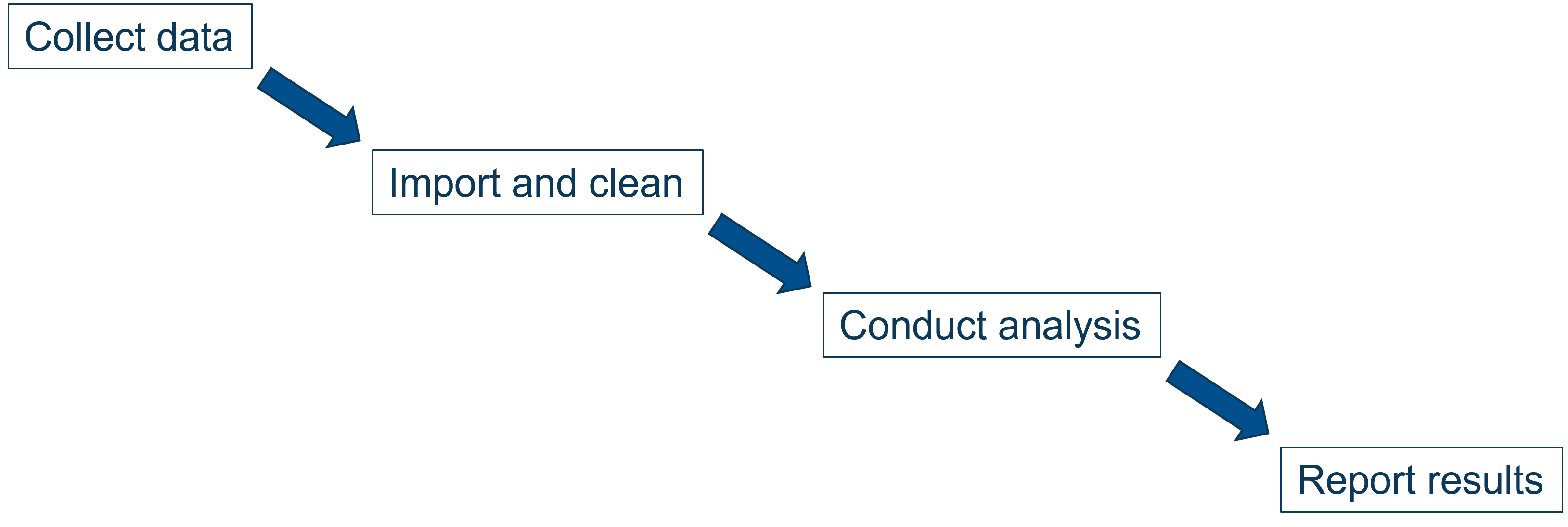

Advantages:
- Reported results are always kept up-to-date with data and analysis
- Changes at any point along the workflow can be made easily and robustly
Data workflow


Advantages:
- Reported results are always kept up-to-date with data and analysis
- Changes at any point along the workflow can be made easily and robustly
R Markdown
Quarto
- We’ll focus on Quarto instead
- Quarto is a relatively new tool very similar to RMarkdown
- Most of what you might read about RMarkdown also applies to Quarto
- see also https://quarto.org for documentation and guides